Advancements in biotechnology have enabled the use of mRNA as a therapeutic to produce nearly all functional proteins in the human body [1]. mRNA-based vaccines and therapeutics hold immense promise in preventing and treating various diseases. Research on mRNA therapeutics is rapidly expanding, and understanding the pharmacodynamics (PD), toxicokinetics (TK), tissue distribution, and pharmacokinetics (PK) of mRNA therapeutics is essential. Accurate quantification of mRNA levels in systemic circulation and tissues is a critical step in ensuring their efficacy and safety.
The RT-qPCR (real-time quantitative PCR) platform is widely regarded as the gold standard for mRNA quantification. Recently, the bDNA (branched-DNA) platform has emerged as a complementary approach to RT-qPCR. In this article, we compare the performance of RT-qPCR and bDNA platforms using a model LNP-mRNA therapeutic in Sprague-Dawley rat plasma, providing insights for researchers conducting mRNA analysis.
Overview of RT-qPCR and bDNA Methods for mRNA Therapeutics Quantification
RT-qPCR method
RT-qPCR is an enzyme-based amplification technique widely used in molecular biology research and diagnostics. The analytical procedures for qualify of mRNA by RT-qPCR include be transcribed into cDNA through the reverse transcription reaction. Subsequently, RT-qPCR amplifies complementary DNA (cDNA) using short complementary primers and probes over multiple thermal cycles, typically ranging from 40 to 45.. SYBR Green dye or TaqMan fluorescent probes are commonly used for detecting amplified DNA products. RT-qPCR methods have been extensively applied in mRNA vaccine and therapeutic research, including the quantification of mRNA encoding antigens in cancer vaccines and mRNA from COVID-19 vaccines (such as mRNA-1273 or BNT162b2) in human plasma and tissues. For example, RT-qPCR has been used to quantify mRNA levels in breast milk and placental explants to evaluate the ability of vaccines to cross the blood-mammary or blood-placental barriers.
bDNA method
The bDNA technique has been used for decades to quantify long nucleic acids and is widely applied in viral load detection for HIV, HCV, and HBV infections. This assay uses custom-designed capture, blocking, and Z-extension probes that hybridize to large regions of the target mRNA sequence. Amplification probes recognize Z-extension probes, and signal amplification is achieved using detection reagents. bDNA assay kits are available in plate-based or bead-based formats. This technology has been used to study the tissue distribution of COVID-19 mRNA vaccines (mRNA-1647) in rats and influenza vaccines encoding hemagglutinin in mice. The principles of both methods are shown in Figure 1.
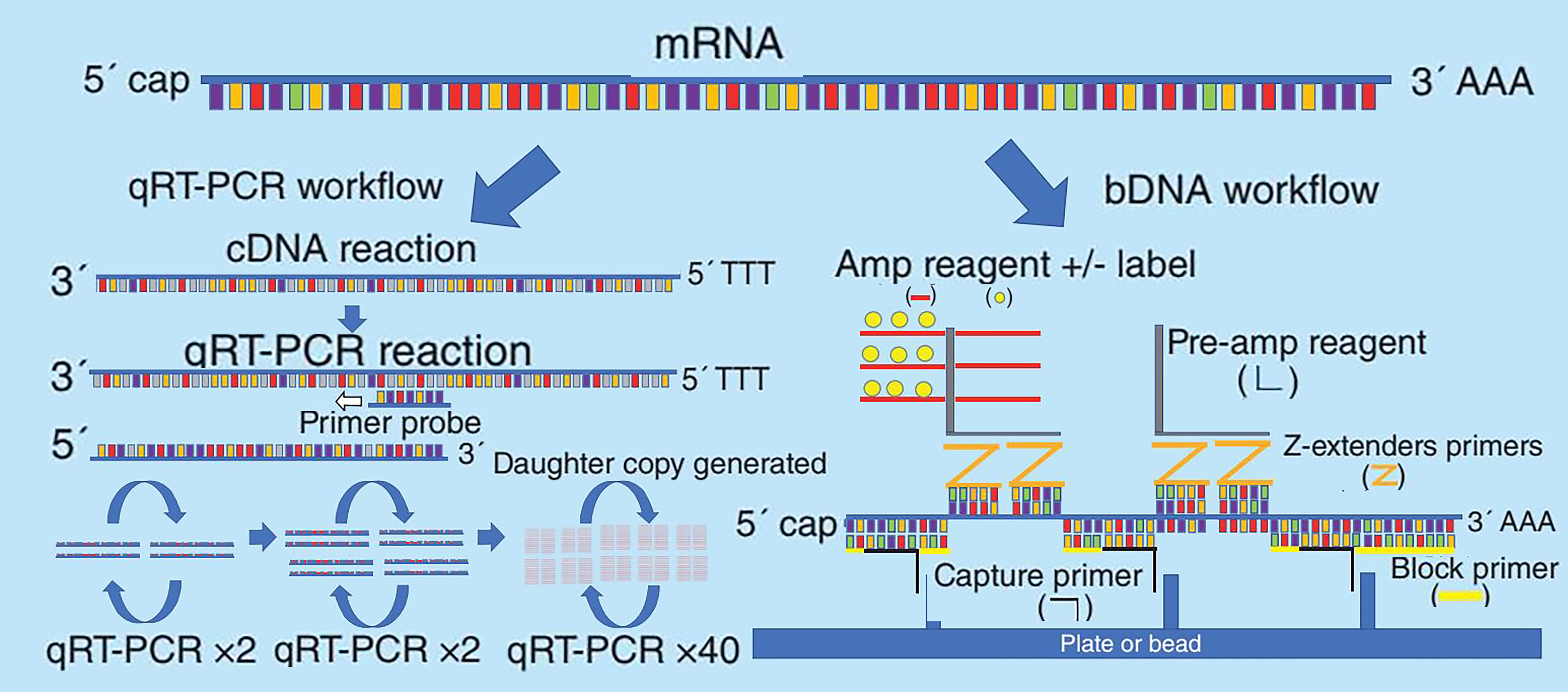
Figure 1. Diagram of RT-qPCR and bDNA method principles [2].
Comparison of RT-qPCR and bDNA mRNA Quantification Performance and Validation Parameters
Workflow comparison
RT-qPCR and bDNA platforms differ in their methodological performance for quantifying LNP-mRNA in plasma. Due to the low endogenous mRNA content in plasma, RT-qPCR uses a direct lysis sample pretreatment, followed by reverse transcription and qPCR steps. bDNA detection employs a modified approach based on methods used for endogenous mRNA quantification. The workflows for the two methods are shown in Figure 2.
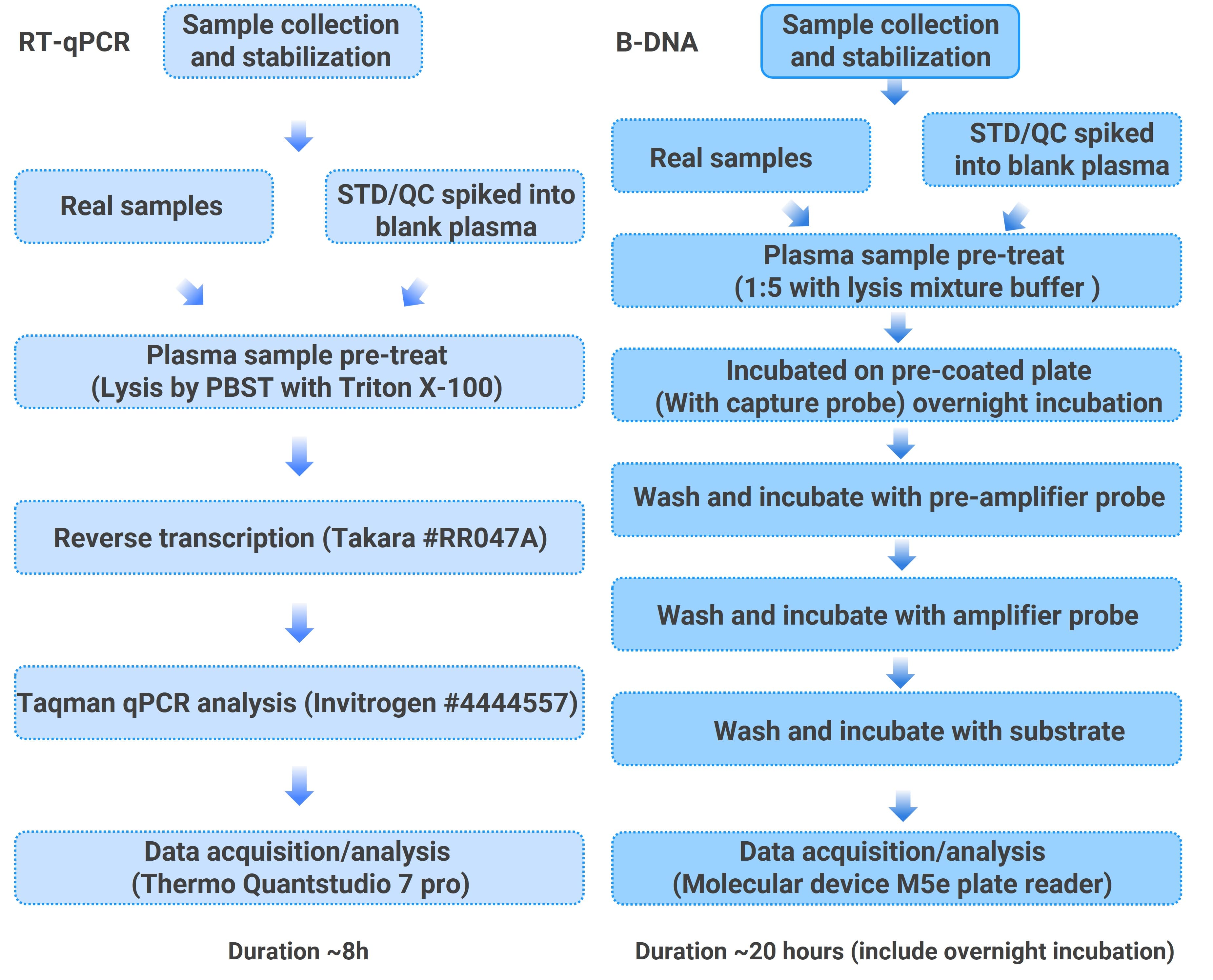
Figure 2. Experimental workflows for quantifying LNP-mRNA (EGFP mRNA) in plasma using RT-qPCR and bDNA methods.
Sensitivity parameters
The lower limit of quantification (LLOQ) for RT-qPCR is 1 pg/mL, in contrast to bDNA, which has an LLOQ of 2 pg/mL. RT-qPCR offers a wider dynamic range spanning from 1 pg/mL to 1 µg/mL, whereas bDNA exhibits a more restricted range of 2–250 pg/mL, as determined by chemiluminescence. The standard curves for both methods are depicted in Figure 3.
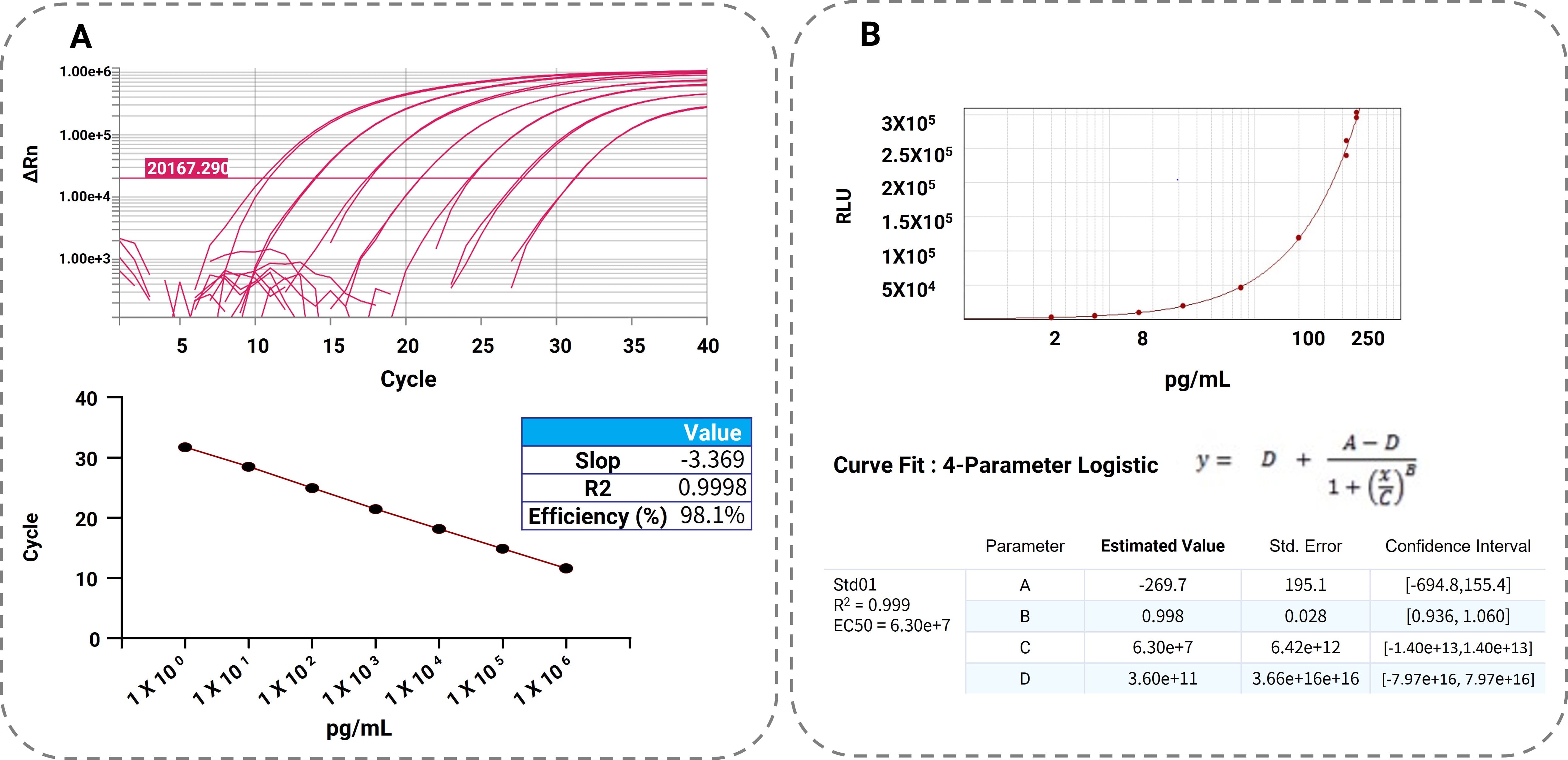
Figure 3A: RT-qPCR amplification and quantification standard curve. The left plot (X-axis: qPCR cycle number; Y-axis: ΔRn) shows standard samples (STD01–STD07) corresponding to concentrations listed in the right plot (X-axis: STD01–STD07 concentrations in pg/mL; Y-axis: qPCR cycle threshold [Ct] values).
Figure 3B: bDNA quantification standard curve (X-axis: STD01–STD08 concentrations in pg/mL; Y-axis: relative light units [RLU]).
Intra-assay and inter-assay accuracy and precision
As shown in Figure 4, each run included one set of standard samples (STD) and three sets of five quality control (QC) samples at different concentrations. For RT-qPCR, the standard curve concentrations were set at 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 1 × 101, and 1 × 100 pg/mL, and QC samples were set at 1 × 106, 5 × 105, 5 × 103, 5 × 101, and 1 × 100 pg/mL. For bDNA, the standard curve concentrations were set at 250, 212.5, 100, 40, 16, 8, 4, and 2 pg/mL, and QC samples were set at 250, 200, 30, 6, and 2 pg/mL.
The bDNA method demonstrated better precision and accuracy overall. The %CV and %Bias for bDNA were generally below 10%, while RT-qPCR showed much higher %CV and %Bias, especially at lower concentrations. For QC samples (ULOQ, HQC, MQC, LQC, LLOQ), the total error (%CV + |%Bias|) for bDNA was 2.8%, 4.4%, 6.5%, 6.7%, and 7.1%, respectively, compared to 19.6%, 22.5%, 19.5%, 30.2%, and 35.6% for RT-qPCR.
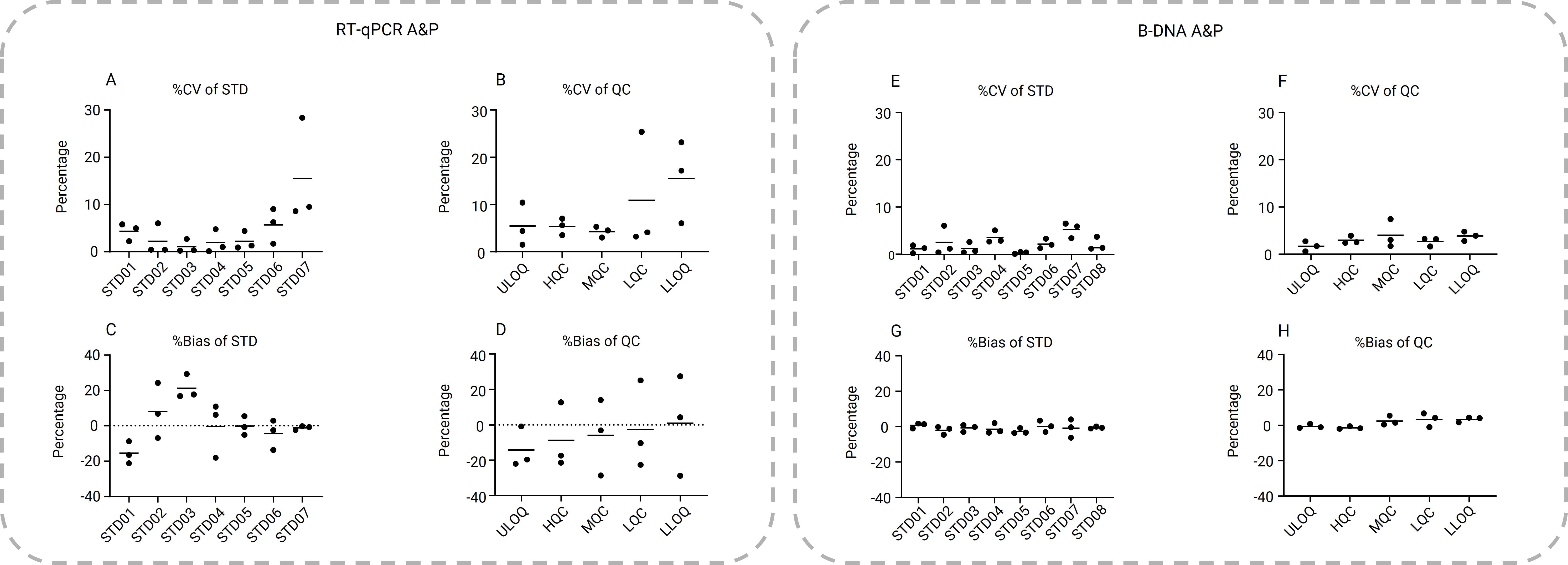
Figure 4. Panels A–D: RT-qPCR intra-assay accuracy and precision (%CV and %Bias) for standard and QC samples across three analytical batches. Panels E–H: bDNA intra-assay accuracy and precision (%CV and %Bias) for standard and QC samples across three analytical batches.
Endogenous mRNA interference tolerance
RT-qPCR demonstrated higher tolerance for endogenous mRNA interference. In experiments where varying concentrations of purified endogenous mRNA from Sprague-Dawley rat liver were artificially added to the reaction systems, RT-qPCR HQC/LQC samples tolerated up to 25 µg/reaction of endogenous mRNA. In contrast, bDNA was less tolerant, with signal suppression observed at 1 µg/reaction, as shown in Figure 5.
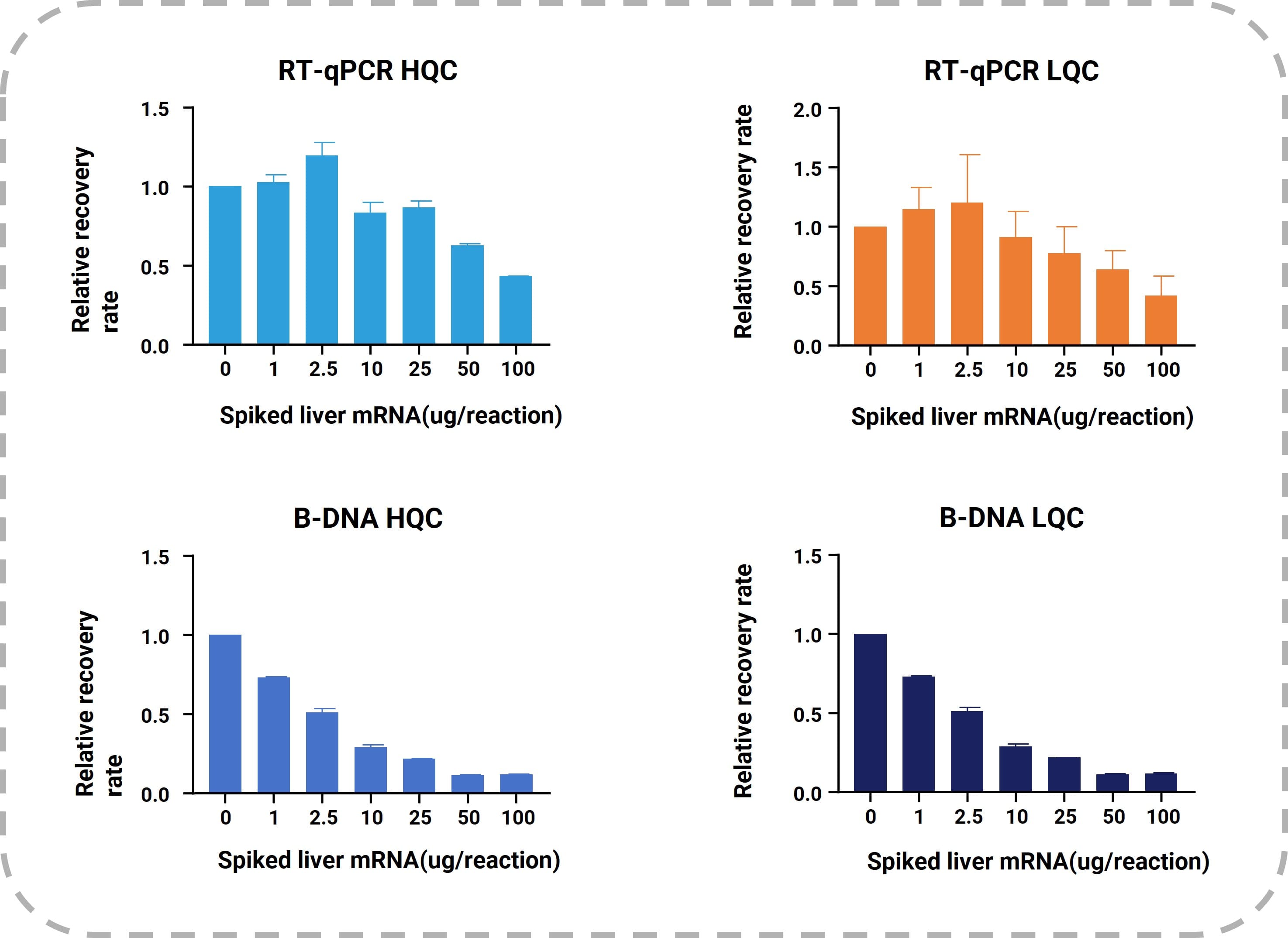
Figure 5. Signal suppression levels in RT-qPCR and bDNA methods caused by high endogenous mRNA concentrations in HQC and LQC samples. X-axis: added endogenous mRNA per reaction; Y-axis: recovery rate (normalized to 1 [100%] for samples without endogenous mRNA).
Selectivity parameters
Both methods exhibited good selectivity in plasma from 10 different male and female Sprague-Dawley rats at various concentrations (ULOQ, HQC, LLOQ). For RT-qPCR, 100% of ULOQ and HQC samples and 94% of the LLOQ samples, met the selectivity criteria. For bDNA, all samples passed the selectivity evaluation.
Stability parameters
Both methods showed good stability for test samples under different conditions, including five freeze-thaw cycles, six hours of room temperature incubation, and 24 hours of incubation at 4°C. The |%Bias| between stability and baseline samples was less than 20% (30% for RT-qPCR).
Summary
This comparison of bDNA and RT-qPCR platforms for LNP-mRNA therapeutics quantification highlights the following:
With optimized methods, bDNA achieves an LLOQ (2 pg/mL) comparable to RT-qPCR (1 pg/mL) but has a narrower quantification range (2–250 pg/mL vs. 1 pg/mL–1 µg/mL for RT-qPCR).
bDNA offers better accuracy and precision, meeting regulatory standards for ligand-binding assay (LBA) platforms.
RT-qPCR is more tolerant of endogenous mRNA interference, making it better suited for tissue samples with high endogenous mRNA variability.
Both methods exhibit comparable selectivity and stability.
Primer/probe design and optimization are critical for both platforms, as proper design significantly enhances detection performance.
Concluding Remarks
mRNA vaccines and therapeutics can theoretically be translated into any protein or peptide via the ribosomal expression system. Compared to DNA-based therapies, mRNA therapeutics do not require nuclear entry, offering higher transfection efficiency and lower toxicity. Importantly, mRNA drugs lack the risks of insertional mutagenesis or infection, reducing concerns about off-target toxicity from non-target DNA modifications. Unlike traditional protein therapies, mRNA therapeutics enable continuous translation of encoded proteins, providing superior efficacy.
The complexity of mRNA therapeutics, including their carriers, expressed proteins, and immunogenic properties, calls for extensive research into mRNA ADME profiles and safety. This necessitates diverse bioanalytical platforms and strategies.
WuXi AppTec DMPK provides preclinical bioanalysis expertise to support mRNA vaccine and therapeutic programs, ensuring efficient and accurate evaluation of ADME profiles and tissue distribution for safety assessments and clinical studies.
Authors: Huan Yan, Miaomiao Song, Maotian Zhou, Xing Lili
Talk to a WuXi AppTec expert today to get the support you need to achieve your drug development goals.
Committed to accelerating drug discovery and development, we offer a full range of discovery screening, preclinical development, clinical drug metabolism, and pharmacokinetic (DMPK) platforms and services. With research facilities in the United States (New Jersey) and China (Shanghai, Suzhou, Nanjing, and Nantong), 1,000+ scientists, and over fifteen years of experience in Investigational New Drug (IND) application, our DMPK team at WuXi AppTec are serving 1,600+ global clients, and have successfully supported 1,700+ IND applications.
Reference
[1] Qin, Shugang, et al. "mRNA-based therapeutics: powerful and versatile tools to combat diseases." Signal transduction and targeted therapy 7.1 (2022): 166.
[2] Henderson, Neil, and Amanda Wilson. "Measurement of mRNA therapeutics: method development and validation challenges." Bioanalysis 11.21 (2019): 2003-2010.
[3] Wissel, Mark, et al. "Recommendations on qPCR/ddPCR assay validation by GCC." Bioanalysis 14.12 (2022): 853-863.
[4] Bioanalytical Method Validation Guidance for Industry, FDA 2018.
Related Services and Platforms




-

 DMPK BioanalysisLearn More
DMPK BioanalysisLearn More -

 Novel Drug Modalities DMPK Enabling PlatformsLearn More
Novel Drug Modalities DMPK Enabling PlatformsLearn More -

 Novel Drug Modalities BioanalysisLearn More
Novel Drug Modalities BioanalysisLearn More -

 Small Molecules BioanalysisLearn More
Small Molecules BioanalysisLearn More -

 Bioanalytical Instrument PlatformLearn More
Bioanalytical Instrument PlatformLearn More -

 PROTAC DMPK ServicesLearn More
PROTAC DMPK ServicesLearn More -

 ADC DMPK ServicesLearn More
ADC DMPK ServicesLearn More -

 Oligo DMPK ServicesLearn More
Oligo DMPK ServicesLearn More -

 PDC DMPK ServicesLearn More
PDC DMPK ServicesLearn More -

 Peptide DMPK ServicesLearn More
Peptide DMPK ServicesLearn More -

 mRNA DMPK ServicesLearn More
mRNA DMPK ServicesLearn More -

 Covalent Drugs DMPK ServicesLearn More
Covalent Drugs DMPK ServicesLearn More
Stay Connected
Keep up with the latest news and insights.