Peptide drugs exhibit significant advantages in targeting "undruggable" targets, such as protein-protein interactions, which are difficult to modulate with traditional small molecule or biologic drugs. Since the first clinical use of insulin in 1921, peptide drugs have evolved significantly, with over 80 approved by regulatory agencies worldwide. Notably, the successful launches of innovative drugs like dulaglutide and semaglutide in recent years has further fueled the research and development of peptide drugs. This article illustrates the focus and challenges in peptide metabolite identification(MetID), introduces an innovative software, PeptideMID, that we developed, and highlights applications through case studies.
Structural characteristics and MetID studies for peptide drugs
Research on peptide drugs began with the basic study of endogenous bioactive substances. Early research focused on elucidating the pharmacological properties of natural human hormones such as insulin, oxytocin, vasopressin, and gonadotropin-releasing hormones. With the breakthrough developments in structural biology, recombinant biotechnology, and novel synthetic and analytical platforms in the 21st century, the development of peptide drugs has been significantly accelerated, establishing a comprehensive and complex research and development system that includes peptide drug discovery, drug design, peptide synthesis, structural modification, and activity evaluation. However, peptide drugs inherently suffer from poor chemical and physiological stability, making them susceptible to proteolytic degradation, resulting in low oral bioavailability, rapid metabolism, and short half-life. Studies have shown that chemical modifications such as cyclization, terminal modifications, and amino acid residue substitutions can significantly enhance peptide stability.
In the drug development process, drug metabolism and pharmacokinetics (DMPK) studies for peptide drugs play a crucial role in ensuring their safety and efficacy. Among these, MetID is a critical research aspect, providing important insights into drug metabolism pathways and assessing potential toxicity risks.
Challenges in peptide MetID and data processing
Peptide drugs play a significant role in treating various diseases, but their chemical and physiological stability is relatively poor, making them prone to proteolytic degradation, resulting in low oral bioavailability, rapid metabolism, and short half-life. To improve their druggability, medicinal chemists employ chemical modifications such as cyclization or bicyclization, N- or C-terminal modifications, substitution of L-amino acids with D-amino acids or non-natural amino acids, and increasing molecular weight, which can significantly enhance peptide stability. However, when non-natural amino acids and/or organic linkers are introduced into peptide structures, new metabolic sites may be generated, necessitating MetID studies to identify metabolic soft spots, continuously optimize molecular structures, and support new drug applications. Challenges in peptide MetID include:
-
Complex metabolic pathways: Peptides have multiple potential cleavage sites, leading to a large number of metabolites. If cyclization is involved, the theoretical number of metabolites can increase exponentially.
-
Difficulty in detection: Peptide drugs and their metabolites usually have low concentrations in vivo and weak UV absorption, making traditional detection methods inadequate and often relying on mass spectrometry data processing and mining.
-
Complex mass spectrometry data interpretation: Peptides form multiply charged ions in mass spectrometry analysis, and side reactions such as dehydration and deamination can occur in tandem with mass spectrometry data, resulting in complex fragment data and increasing the difficulty of fragment ion assignment.
Currently, data processing remains a rate-limiting step in peptide MetID research. Existing technical approaches mainly rely on background subtraction, manual interpretation, and limited commercial software assistance.
Many existing tools struggle with the complexity of peptide structures, particularly when it comes to cyclic peptides or those modified with non-natural amino acids. This can lead to a high incidence of false positives, as the software may not adequately recognize or interpret these variations. Furthermore, while some tools are optimized for small molecule analysis, they may lack the necessary precision when applied to larger molecules or complex peptide modifications.
Applications and case studies of software PeptideMID
To address the technical challenges of MetID for complex peptide structures and to overcome the limitations of commercial software, our team successfully developed a platform for peptide drug MetID, PeptideMID, which includes the PeptideID and PeptideMS2 modules. The PeptideID module can automatically predict metabolites based on peptide amino acid sequences and structural modifications and construct a mass spectrometry database. It supports various backbone types, including linear peptides, cyclic peptides, multi-cyclic peptides, and stapled peptides, as well as custom input of non-natural amino acids and organic groups. By automatically matching high-resolution mass spectrometry data, it quickly identifies potential metabolites. It optimizes matching for complex structures, overcoming the limitations of traditional software. The PeptideMS2 module is specifically designed to interpret peptide tandem mass spectrometry data, automatically identifying key fragment ions and improving the accuracy of MetID studies. It is suitable for complex peptide structures, enabling high-throughput data processing. We demonstrate the applicability and effectiveness of our in-house developed software through several case studies.
Case study 1: In vitro kidney S9 MetID study of three marketed peptides
Three marketed peptide drugs, Leuprorelin, Cetrorelix, and Desmopressin, were incubated in kidney S9 for 90 minutes and analyzed using LC-HRMS. Figure 1 compares the metabolites identified by PeptideMID with those reported in the literature. The results show that the PeptideMID platform effectively identifies and interprets metabolites, demonstrating advantages in efficiency and quality compared to the literature-reported method combining CD software and manual interpretation.
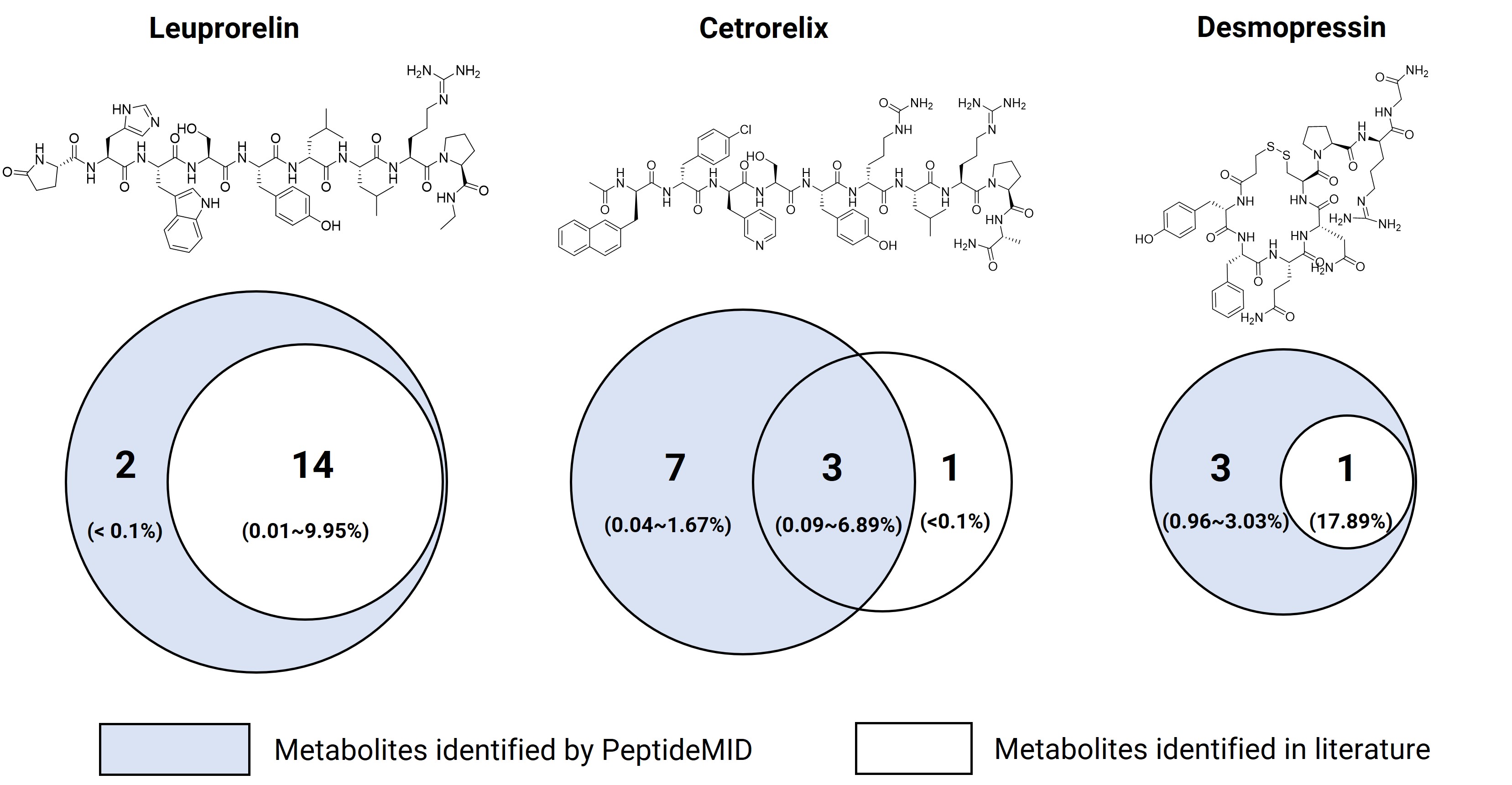
Figure 1. Comparison of in vitro kidney S9 metabolite identification results for three marketed peptides using PeptideMID and literature reports
Case study 2: In vitro plasma metabolite identification study of Peptide-1
After incubating Peptide-1 in vitro in plasma, LC-HRMS was used for detection. As shown in Figure 2, PeptideMID successfully detected four metabolites, M1-M4. M1, M2, and M4 are isomers, all products of a single peptide bond hydrolysis of Peptide-1, with M4 having a relative abundance exceeding 30%. By analyzing the mass spectrometry fragments, the hydrolysis site of M4 was precisely located at the peptide bond between the eighth and ninth amino acids. The results indicate that PeptideMID effectively identifies and interprets peptide metabolites, providing robust scientific evidence and data support for peptide structure optimization and drug development.
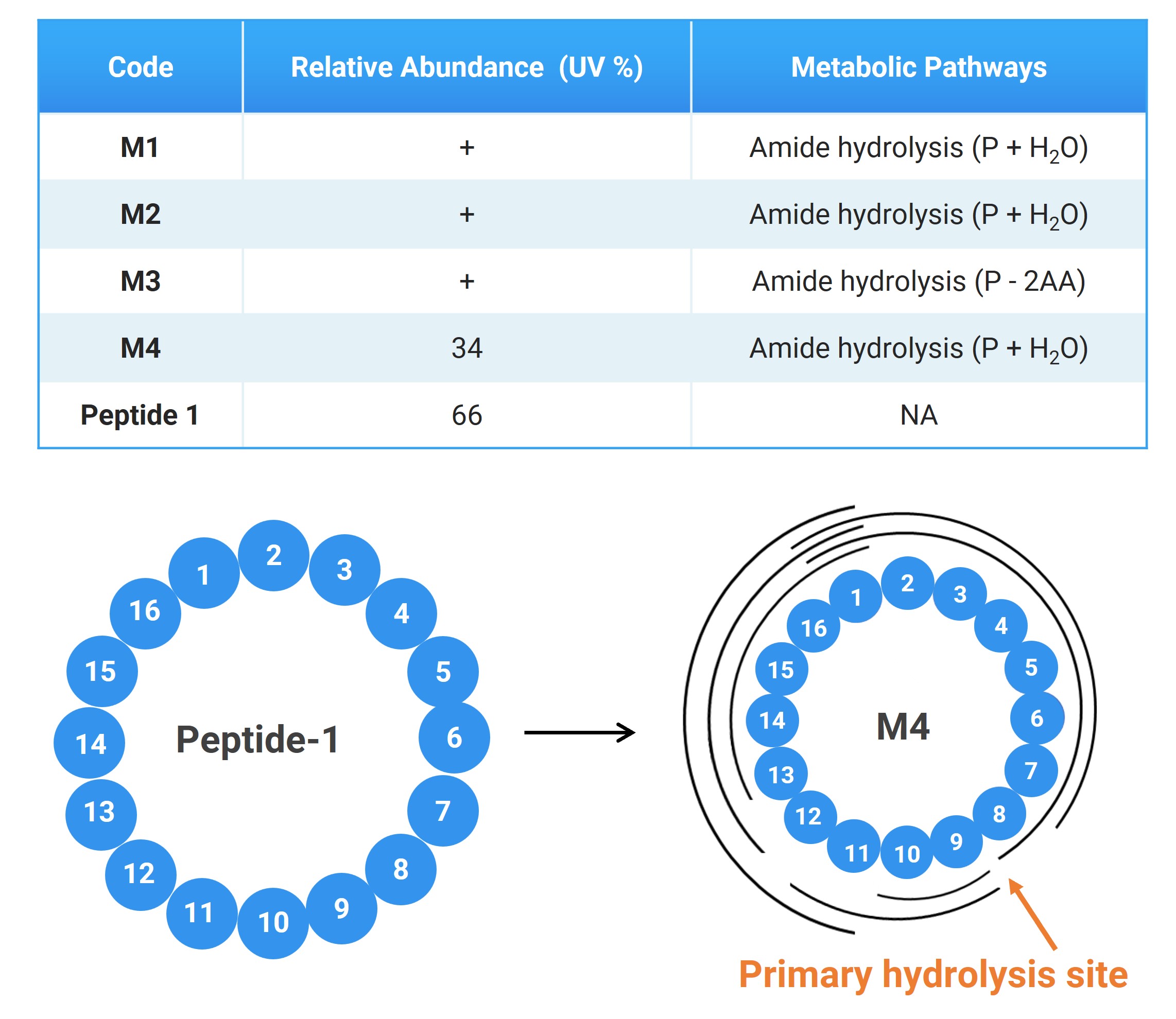
Figure 2. In vitro plasma metabolite study results for Peptide-1 and fragment analysis of M4
Concluding remarks
Peptide drugs play a significant role in treating cancer, immunomodulation, and metabolic diseases. However, their inherent instability (e.g., susceptibility to proteolytic degradation) leads to pharmacokinetic challenges such as low oral bioavailability and short plasma half-life, affecting clinical translation. Chemical modifications such as cyclization, terminal modifications, and amino acid substitutions can significantly enhance peptide stability. However, the increasing complexity of peptide structures poses challenges for metabolite identification.
Metabolite identification research is crucial throughout the drug development cycle: identifying metabolic soft spots in the non-clinical stage to guide lead compound optimization and support clinical applications; in the clinical stage, focusing on human-specific metabolites or disproportionate metabolites (>10% of total drug-related exposure) to guide safety studies. WuXi AppTec DMPK has successfully developed and implemented the metabolite analysis software, PeptideID and PeptideMS2, improving the efficiency and accuracy of MetID research. The advancements brought by PeptideID and PeptideMS2 represent an important leap forward in metabolite identification. These technologies not only enhance the precision and reliability of MetID research but also contribute to the overall success of peptide drug development, facilitating the delivery of new treatments to patients in a more efficient and safe manner.
Authors: Peng Li, Weiqun Cao
Talk to a WuXi AppTec expert today to get the support you need to achieve your drug development goals.
Committed to accelerating drug discovery and development, we offer a full range of discovery screening, preclinical development, clinical drug metabolism, and pharmacokinetic (DMPK) platforms and services. With research facilities in the United States (New Jersey) and China (Shanghai, Suzhou, Nanjing, and Nantong), 1,000+ scientists, and over fifteen years of experience in Investigational New Drug (IND) application, our DMPK team at WuXi AppTec are serving 1,600+ global clients, and have successfully supported 1,700+ IND applications.
Reference
[1] He M M, Zhu S X, Cannon J R, et al. Metabolism and excretion of therapeutic peptides: Current industry practices, perspectives, and recommendations[J]. Drug Metabolism and Disposition, 2023, 51(11): 1436-1450.
[2] Yao J F, Yang H, Zhao Y Z, et al. Metabolism of peptide drugs and strategies to improve their metabolic stability[J]. Current Drug Metabolism, 2018, 19(11): 892-901.
[3] Jyrkäs J, Lassila T, Tolonen A. Extrahepatic in vitro metabolism of peptides; comparison of human kidney and intestinal S9 fraction, human plasma and proximal tubule cells, using cyclosporine A, leuprorelin, and cetrorelix as model compounds[J]. Journal of Pharmaceutical and Biomedical Analysis, 2023, 225: 115219.
Related Services and Platforms




-

 MetID (Metabolite Profiling and Identification)Learn More
MetID (Metabolite Profiling and Identification)Learn More -

 Novel Drug Modalities DMPK Enabling PlatformsLearn More
Novel Drug Modalities DMPK Enabling PlatformsLearn More -

 In Vitro MetID (Metabolite Profiling and Identification)Learn More
In Vitro MetID (Metabolite Profiling and Identification)Learn More -

 In Vivo MetID (Metabolite Profiling and Identification)Learn More
In Vivo MetID (Metabolite Profiling and Identification)Learn More -

 Metabolite Biosynthesis and Structural CharacterizationLearn More
Metabolite Biosynthesis and Structural CharacterizationLearn More -

 Metabolites in Safety Testing (MIST)Learn More
Metabolites in Safety Testing (MIST)Learn More -

 PROTAC DMPK ServicesLearn More
PROTAC DMPK ServicesLearn More -

 ADC DMPK ServicesLearn More
ADC DMPK ServicesLearn More -

 Oligo DMPK ServicesLearn More
Oligo DMPK ServicesLearn More -

 PDC DMPK ServicesLearn More
PDC DMPK ServicesLearn More -

 Peptide DMPK ServicesLearn More
Peptide DMPK ServicesLearn More -

 mRNA DMPK ServicesLearn More
mRNA DMPK ServicesLearn More -

 Covalent Drugs DMPK ServicesLearn More
Covalent Drugs DMPK ServicesLearn More
Stay Connected
Keep up with the latest news and insights.